Antibody Variable Domain Structures in the PDB393 Files containing antibody structures were extraced from the PDB database and dissected into individual domains, yielding 407 VH, 392 Vk and 111 Vl domains (including multiple copies in the assymetric unit, but excluding the multiple models in NMR files). These represent 203 nonredundant VH domains, 195 nonredundant Vk and 30 nonredundandant Vl domains (domains with distinct sequences). The residue numbers were changed to the AHo nomenclature to facilitate aufomated processing. The renumbered files can be downloaded from this site, either individually or as a compressed tar archive. |
The results presented in this part of the website were derived from the analysis of these domain structures. They include the analysis of the sequence and structural variability (deviation from the average structure), the side chain solvent accessibility, the residues involved in antigen contact and in VH/VL dimer contact, the torsion angles and conserved hydrogen bonding pattern. Things to come: A similar analysis of the TCR variable domains will be added in the near future. |
||||||
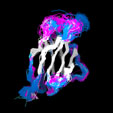
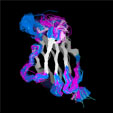
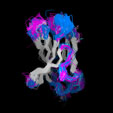
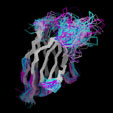
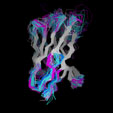
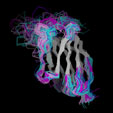
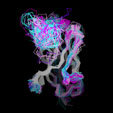
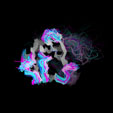
Different Views of the 3D-aligned VL-domains:(pink: Vl, L7 or L9 not Pro, magenta: Vl, L7 and L9 = Pro, cyan: Vk, L8 not Pro, blue: Vk, L8 = cis-Pro). Click on any of the images for an enlarged view
|
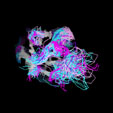
Different Views of the 3D-aligned VH-domains:(magenta: Type I, H6 Glu, H7 not Pro, H10 Pro; pink: Type II, H6 Glu, H7 not Pro, H10 Gly; cyan: Type III, H6 Gln, H7 not Pro; blue: Type IV, H6 Gln, H7 Pro) |
|
|
|||||||||||||||||
|
Last Modified by A.Honegger |
|||||||||||||||||