HuCAL VLl Germline Consensus Sequences
click on any of the shaded areas to see an alignment of the germline sequences represented by the cluster to the corresponding HuCAL consensus sequence |
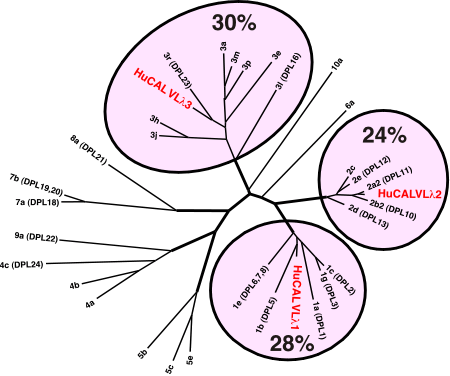
Coverage of human VL lambda germline sequence space by HuCAL sequencesThe protein sequences representing the human VL and VH germlines were taken from VBase and aligned to the 14 HuCAL sequences. The Phylip and ClustalW (Higgins et al., 1994 phylogeny program packages were used to generate separate rootless trees for the VL kappa, VL lambda and VH sequences. Percentages indicate the fraction of rearranged sequences in the Kabat database which cluster with the different germline subgroups. For this calculations, we used a database of rearranged sequences with 937 VL kappa, 421 VL lambda and 1267 VH sequence entries. |
|
|
|||||||||||||||||
|
Last Modified by A.Honegger |
|||||||||||||||||