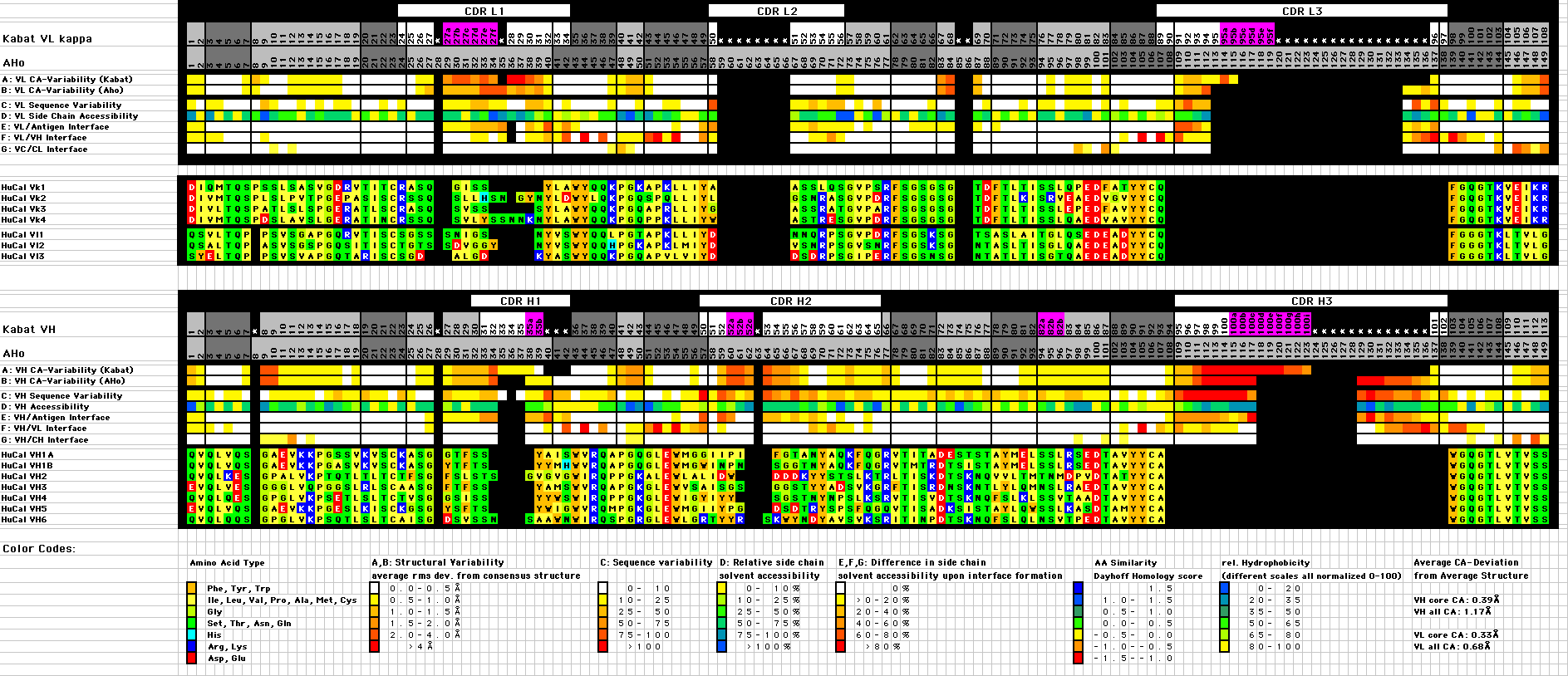
| (A,B,C) Structural variability: Average rms deviation from mean Ca position (Average of 185 Vk and 206 VH structures representing >100 non-identical sequences, all the experimental Fv and Fab structures with a resolution better than 3.0Å available in the PDB database at the time of the analysis) for structures aligned according to the AHo (A), Chotia (B) and Kabat (C) and alignment. Individual domains were excised from the corresponding PDB files and aligned by a least squares fit of the Ca-positions of the core residues (3-7, 20-24, 41-47, 51-57, 78-82, 89-93, 102-108 and 138-144) to the corresponding Ca-positions of a reference structure 1YEH (Gigant et al., 1997) for Vk and 1MFD (Bundle et al., 1994) for VH. The mean Ca positions for each residue were calculated and the average deviation for each residue position in the alignment is indicated by a color code (white: rms deviation < 0.5 Å, yellow: 0.5-1 Å, yellow-orange: 1-1.5 Å, orange 1.5-2 Å, orange-red 2-4 Å, red >4 Å) |
(D) Sequence variability: From the position-dependent amino acid composition, the sequence variability was calculated according to Kabat et al. (1991): Variability = 100*(number of different amino acids at position n) / (frequency of the most common amino acid at that position). Color code: white: <10, yellow: 10-25, yellow-orange: 25-50, orange: 50-75, red-orange: 75-100, red: >100 |
(E) Average relative side chain accessibility: The side chain solvent accessible surface of each residue was calculated as percentage of the solvent accessible surface the same residue would have in the context of a poly-Ala peptide in extended conformation (program NACCESS (http://sjh.bi.umist.ac.uk/naccess.html)) (yellow: 0-10%, yellow-green: 10-25%, buried; green: 25-50%, green-blue: 50-75%, semi-buried; blue: 75-100%, dark blue: >100%, exposed). |
(F) Reduction of the side chain accessible surface upon formation of the complex of the Fv fragment with a protein antigen: Relative reduction of the side chain accessible surface of each residue in the complex of antigen-Fv fragment compared to the same residue in the Fv fragment without antigen (white: 0% reduction, yellow: 0-20%, yellow-orange: 20-40%, orange: 40-60%, red-orange: 60-80%, red: 80-100%). |
(G) Reduction of the side chain accessible surface upon formation of the dimer interface between VL and VH: Average relative reduction of the side chain accessible surface of each residue in the Fv fragment compared to its accessible surface in the isolated VL or VH domain (white: 0% reduction, yellow: 0-20%, yellow-orange: 20-40%, orange: 40-60%, red-orange: 60-80%, red: 80-100%). |
(H) Reduction of the side chain accessible surface upon formation of the interface between VL and CL or between VH and CH: Average relative reduction of the side chain accessible surface of each residue in the Fab fragment compared to its accessible surface in the Fv fragment (white: 0% reduction, yellow: 0-20%, yellow-orange: 20-40%, orange: 40-60%, red-orange: 60-80%, red: 80-100%). |
![]() Numbering Schemes
Numbering Schemes ![]() Alignment of reference sequences
Alignment of reference sequences ![]() Deviation from average structure
Deviation from average structure