 |
- Antibody Vl Framework 1
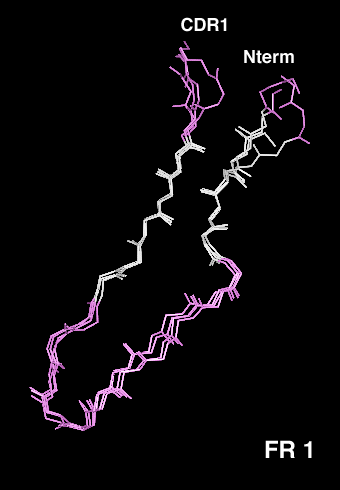
- The b-sandwich structure of immunoglobulin variable domains is characterized by a typical kink in the first strand, which allows the first part of the strand to hydrogen bond to the outer beta-sheet (away from the VH-VL interface) and the second part to the inner beta-sheet. This kink differs in length and sequence between the Vk, Vl and VH domains and yet is involved in several almost perfectly conserved interactions with framework residues. In Vk domains, a cis-Pro is frequently found in position L8 (blue). In Vk domains which do not contain a Pro in this position, the corresponding amino acid shows a normal trans peptide bond, and the main chain conformation has to adapt accordingly (cyan). Vl domains show a one residue deletion (D L8) compared to Vk (magenta).
-
- We have used the selectively infective phage (SIP) system to select the optimal kink region from several defined libraries, using an anti-hemagglutinin single-chain Fv (scFv) fragment as a model system. Both for the kink with the Vk domain length and that with the Vl length, a sequence distribution was selected that coincides remarkably well with the sequence distribution of natural antibodies. The selected scFv fragments were purified and characterized, and thermodynamic stability was found to be the prime factor responsible for selection. The VL domains with kappa-like length and a proline residue in position L8 were found to be the most stable of the mutants tested.
-
-
- Spada, S., Honegger, A. and Plückthun, A., Reproducing the natural evolution of protein structural features with the selectively infective phage (SIP) technology. The kink in the first strand of antibody kappa domains, J. Mol.Biol. 283, 395-407 (1998)

|