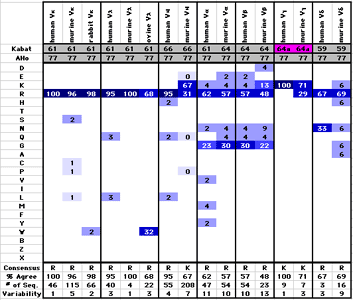
Sequence StatisticsThe sequence statistics macros perform some common sequence statistics tasks on a sequences alignment: They calculate the amino acid composition of the sequences, count the occurance of different amino acids at given positions in a sequence alignment, determine the consensus sequence, sequence variability and frequency of the most abundant amino acid, and convert the numeric values generated by these analyses to color codes. Example: The coloring of the frequency table makes it immediately apparent that this particular position (Residue 77) shows a strong preference for basic residues in all types of IG variable domains
|
Macros: AA_SeqLength counts number of valid amino acid within the selected sequences. The result is placed in the column following the selected sequence block. AA_Composition determines the amino acid composition of all sequences in an alignment AA_Frequency determines the frequency of different amino acids in each position of a sequence alignment, the consensus sequence, the percentage of sequences which agree with the consensus and the Kabat sequence variability at this position. AA_FreqValColor colors the table generated by AA_Frequency by valueas shown on the left AA_SeqVarColor converts the Kabat sequence variability values to a color code AA_FreqDistribute rearranges a series of tables generated by AA_Frequency in such a way that the results for each residue position are collected in a separate table on a separate worksheet AA_Similarity colors a sequence alignment according to similarity to a reference sequence. Calculated %sequence similarity, % sequence identity and the length of each sequence (color code) AA_Differences Colors a sequence alignment in such a way that the differences between neighboring sequences are highlighted and counts the number of differences between a sequence and the next one in the alignment |
|
|
|||||||||||||||||
|
Last Modified by A.Honegger |
|||||||||||||||||