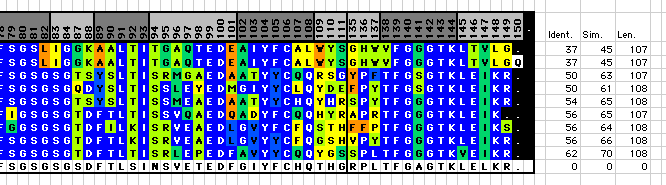
Macro AA_SimilarityColors a sequence alignment according to similarity to the reference sequence (the last sequence in the selected sequence block). The degree of similarity is taken from the Dayhof PAM matrix (worksheet "Hom.Score" in workbook "AHo_macros.xls")
The overall % sequence identity, % sequence similarity and sequence length are indicated to the right of the sequence alignment. Color Code: dark blue=identical, blue: very conservative substitution, blue-green: conservative substitution, yellow-green: somewhat similar, yellow and orange: increasingly different. Usage:
Caution: Four cells to the right of the sequence block will be overwritten with the %Identity, %Similarity and Sequence Length results
Sub AA_Similarity()
'colors amino acid sequences by identity/similarity
'Assumption: The last sequence in the selection is the reference sequence
If Selection.Columns.Count = 0 Then 'Error, nothing selected
MsgBox Prompt:="No cells selected, please select the sequences you wish to color"
Exit Sub
End If
'get extent of current selection
i1 = Selection.Row
i2 = i1 + Selection.Rows.Count - 1
j1 = Selection.Column
j2 = j1 + Selection.Columns.Count - 1
For i = i1 To i2 Step 1
Cells(i, j2 + 2) = 0 'identity
Cells(i, j2 + 3) = 0 'similarity
Cells(i, j2 + 4) = 0 'length
Next i
'Format selected area
Selection.Borders(xlLeft).LineStyle = xlNone
Selection.Borders(xlRight).LineStyle = xlNone
Selection.Borders(xlTop).LineStyle = xlNone
Selection.Borders(xlBottom).LineStyle = xlNone
Selection.BorderAround Weight:=xlThick
Selection.Interior.ColorIndex = xlNone
Selection.Font.Name = "Geneva"
Selection.Font.FontStyle = "Regular"
Selection.Font.Size = 9
Selection.Font.ColorIndex = 1
Selection.Font.Bold = True
Selection.HorizontalAlignment = xlCenter
Selection.VerticalAlignment = xlCenter
For j = j1 To j2 Step 1 'column j1-j2'
For k = 2 To 23 Step 1 'identify reference amino acid'
If (Cells(i2, j) = Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, 1)) Then
Exit For
End If
Next k
o = 0
p = 0
For i = i1 To i2 - 1 Step 1 'row i1-(i2-1)'
For l = 2 To 23 Step 1 'identify amino acid'
If (Cells(i, j) = Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(1, l)) Then
Exit For
End If
Next l
N = 3
If (l > 23) Then
N = 1
ElseIf (k > 23) Then
N = 2
Else
Cells(i, j2 + 4) = Cells(i, j2 + 4) + 1
Cells(i, j2 + 3) = Cells(i, j2 + 3) + (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) / 1.5)
If (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) = 1.5) Then
N = 43
Cells(i, j2 + 2) = Cells(i, j2 + 2) + 1
ElseIf (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) > 1) Then N = 42
ElseIf (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) > 0.5) Then N = 40
ElseIf (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) > 0) Then N = 38
ElseIf (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) > -0.5) Then N = 37
ElseIf (Workbooks("AHo_macros.xls").Sheets("Hom.Score").Cells(k, l) > -1) Then N = 35
Else: N = 33
End If
End If
Cells(i, j).Select
With Selection.Interior
.ColorIndex = N
.Pattern = xlSolid
If N = 43 Or N = 33 Or N = 1 Then
Selection.Font.ColorIndex = 2
End If
End With
Next i
Next j
For i = i1 To (i2 - 1) Step 1
If (Cells(i, j2 + 4) > 0) Then
Cells(i, j2 + 2) = (100 * Cells(i, j2 + 2)) / Cells(i, j2 + 4) '% identity
Cells(i, j2 + 3) = (100 * Cells(i, j2 + 3)) / Cells(i, j2 + 4) '% similarity
End If
Next i
Range(Cells(i1, j2 + 1), Cells(i2, j2 + 1)).Select
Selection.ColumnWidth = 1
Range(Cells(i1, j2 + 2), Cells(i2, j2 + 4)).Select
Selection.ColumnWidth = 5
Selection.NumberFormat = "0"
Cells(i1 - 1, j2 + 2) = "Ident."
Cells(i1 - 1, j2 + 3) = "Sim."
Cells(i1 - 1, j2 + 4) = "Len."
End Sub
|
|
|
|||||||||||||||||
|
Last Modified by A.Honegger |
|||||||||||||||||