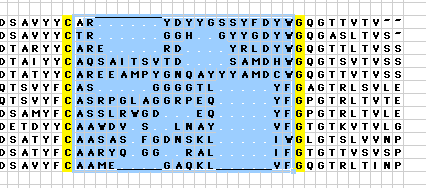
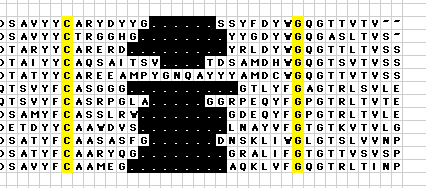
Macro AA_CDR_ArrangeThe AHo numbering scheme specifies that sequence gaps should be centered around the turn position in the longest sequence variant, or centered between corresponding positions in neighboring beta-strands. The macro AA_CDR_arrange helps to align sequences in this manner, since it coalesces all gaps in a selected sequence block in the center of this block: Usage: To coalesce all the gaps in the center between the highlighted Cys and the highlighted Gly Sub AA_CDR_Arrange()
'Counts number of valid amino acid within selected rows
'Consolidates gaps and places them in the center of the sequence block
If Selection.Columns.Count = 0 Then 'Error, nothing selected
MsgBox Prompt:="No cells selected, please select the sequences you wish to color"
Exit Sub
End If
'get extent of current selection
i1 = Selection.Row
i2 = i1 + Selection.Rows.Count - 1
j1 = Selection.Column
j2 = j1 + Selection.Columns.Count - 1
For i = i1 To i2 Step 1
a = 0
b = 0
For j = j2 To j1 Step -1
If (Cells(i, j) = "" Or Cells(i, j) = " ") Then
Cells(i, j) = "."
End If
If (Cells(i, j) = ".") Then
Cells(i, j).Select
Selection.Delete Shift:=xlToLeft
a = a + 1
Else
b = b + 1
End If
Next j
If Not (a = 0) Then
Range(Cells(i, j1 + Int((b + 0.99) / 2)), Cells(i, j2 - Int((b - 1) / 2) - 1)).Select
Selection.Insert Shift:=xlToRight
End If
Next i
Range(Cells(i1,j1), Cells(i2,j2)).Select
End Sub
|
|
|
|||||||||||||||||
|
Last Modified by A.Honegger |
|||||||||||||||||