|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Designer Proteins
Designer Proteins
As we learn more and more about proteins and how they work, we naturally have the desire to use this knowledge and do some tinkering of our own. Since the early 1980's, scientists have been using the ever-expanding understanding of protein structure and function to redesign existing proteins, and more recently, to design entirely new proteins.
Designing Proteins
As scientists began this quest, they quickly found that proteins are more complicated than they might seem. The different types of amino acids, each with their own chemical features, work together to coax a protein chain to fold into a compact stable structure. A collection of carbon-rich amino acids, like leucine and phenylalanine, are usually placed inside the protein, all chosen to lock perfectly together. On the other hand, charged amino acids, such as lysine and aspartic acid, are typically spread across the surface to make the protein soluble in water. Hydrogen-bonding amino acids, such as serine and asparagine, are dotted in strategic places to tie different portions of the chain together. Finally, the odd glycine or proline is added to redirect the chain in the proper direction.Negative Design
This combination of favorable forces locks the protein chain into a stable, compact structure. But this is only the first step in protein design. In order to design a protein that will successfully fold, you also need to make sure that the protein only has one stable structure. If there are any other ways to fold the protein, they will compete with your desired structure and spoil the construction. So, it is not enough design a stable protein structure...you also need to design a protein that is unstable in every other conformation.Proteins from Scratch
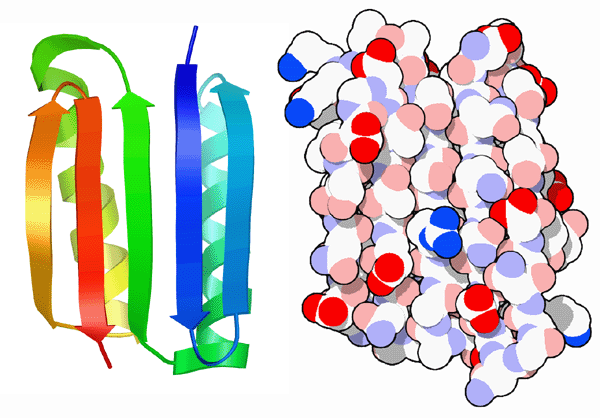
Building on decades of protein structure research, scientists at the Fred Hutchinson Cancer Research Center and the University of Washington in Seattle have successfully weighed these positive and negative design elements to create an entirely novel protein. They started with a fold that had never been seen in nature, shown here on the left. Then, they used a computational method to design a sequence of amino acids that would adopt this fold. When they built this protein, which they call Top7, they found that it folded up and adopted exactly the structure they designed, as seen in PDB entry 1qys.Next: Amazing Alpha Helices
Last changed by: A.Honegger,