|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
p53 Tumor Suppressor
Exploring the Structure
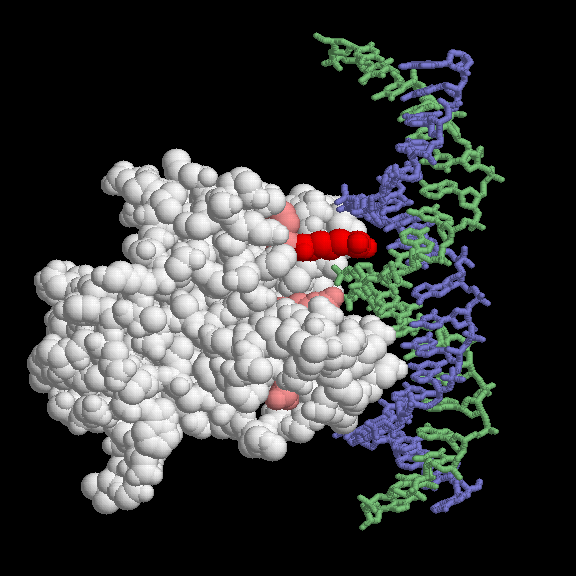
Most of the p53 mutations that cause cancer are found in the DNA-binding domain. The most common mutations are shown here, using PDB entry 1tup. This PDB entry includes three copies of the DNA-binding domain; only one (chain B in the file) is shown here. The mutations are found in and around the DNA-binding face of the protein. The most common mutation changes arginine 248, colored red here. Notice how it snakes into the minor groove of the DNA (shown in blue and green), forming a strong stabilizing interaction. When mutated to another amino acid, this interaction is lost. Other key sites of mutation are shown in pink, including arginine residues 175, 249, 273 and 282, and glycine 245. Some of these contact the DNA directly, and others are involved in positioning other DNA-binding amino acids.This picture was created with RasMol. You can create similar illustrations by clicking on the accession codes and picking an option under View Structure.
A list of all p53 tumor suppressor structures in the PDB as of July, 2002, is available here. For more information on p53 tumor suppressor, click here.
Next: Interaktive 3D-Animation
Previous: Embracing DNA
Last changed by: A.Honegger,