|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Aminoacyl-tRNA Synthetase
Exploring the Structure
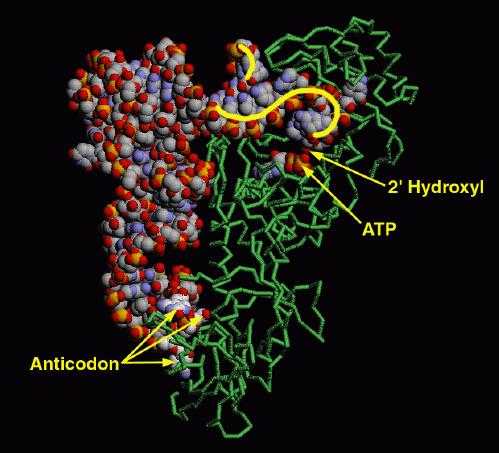
These enzymes are not gentle with tRNA molecules. The structure of glutaminyl-tRNA synthetase with its tRNA (entry 1gtr) is a good example. The enzyme firmly grips the anticodon, spreading the three bases widely apart for better recognition. At the other end, the enzyme unpairs one base at the beginning of the chain, seen curving upward here, and kinks the long acceptor end of the chain into a tight hairpin, seen here curving downward. This places the 2' hydroxyl on the last nucleotide in the active site, where ATP and the amino acid (not present in this structure) are bound.
This illustration was created with RasMol, using a backbone representation for the protein (chain A) and spacefilling representations for the tRNA chain (chain B) and the ATP molecule (residue name ATP). You can create similar pictures by clicking on the accession code above, and then picking one of the options under "View Structure". Try looking also at the many protein sidechains that interact with the tRNA.
A list of Aminoacyl-tRNA Synthetases in the PDB as of April, 2001, is available here. For suggestions for further information on aminoacyl-tRNA synthetases, click here.
Next: Interaktive 3D-Animation
Previous: High Fidelity
Last changed by: A.Honegger,