|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Zinc Fingers
As you are browsing through the proteins in the PDB, you may notice something: most proteins are big. They contain hundreds of amino acids, even though most of the work is often done by a few amino acids on one side. Why are proteins so big? One reason that proteins are so large is that they must self-assemble inside cells. Proteins are built as floppy chains that fold all by themselves (or with a little help from chaperones) into stable, compact structures. These folded structures are stabilized by hydrogen bonds, charge-charge interactions and hydrophobic forces between the different amino acids, which all line up like pieces in a jigsaw puzzle when the protein folds. A single hydrogen bond or a few charge pairs would not be enough, but a chain of hundreds of amino acids has hundreds of interactions that together glue the protein into a stable structure.
Small and Mighty
Our cells, on the other hand, often use a zinc atom to take a short cut. By arranging two cysteines and two histidines close to each other in a chain, a protein can grab a zinc ion and fold tightly around it. In these proteins, termed zinc fingers, a short chain of 20-30 amino acids is enough to create a solid, stable structure. Zinc fingers are so useful that they are found in thousands of our proteins, and are common in all plants and animals. Surprisingly, however, bacteria do not appear to take advantage of these little structures.
Sticky Fingers
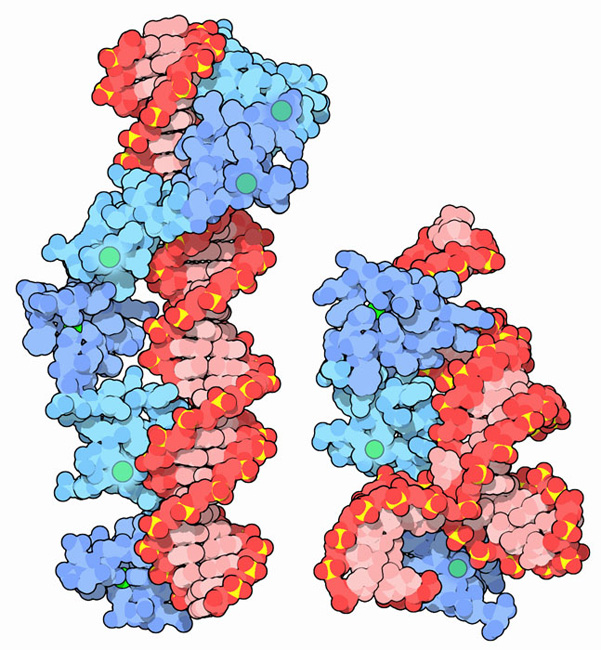
Many zinc fingers play essential roles in DNA recognition. Zinc fingers were originally discovered in the transcription factor TFIIIA from frog eggs, which contains nine zinc fingers in a row. The two structures shown here capture pieces of this protein performing its functions. PDB entry 1tf6, shown on the left, includes 6 of the zinc fingers (blue) bound to a long stretch of DNA (red). With this interaction, TFIIIA helps control the transcription of the gene for a ribosomal RNA. TFIIIA also binds to the ribosomal RNA itself, as shown in PDB entries 1un6 and 2hgh, on the right. These structures include 3 of the 9 zinc fingers (blue), along with a small piece of the ribosomal RNA (red). In the frog eggs, about 10 billion copies of this protein help to stabilize the many copies of RNA as the cell develops.
Modular Recognition
As you can see from these structures, the string of zinc fingers curls along the DNA or RNA strands, binding in the grooves and extending amino acids inwards to read the bases. A single zinc finger does not bind very tightly and can only recognize 2 or 3 base pairs. But when several are strung together, the group binds more tightly and can read longer DNA sequences. This modular approach is so appealing that researchers are currently trying to design artificial zinc fingers with different specificities. Then, by linking them up in the proper order, we could create a custom zinc finger protein to read any sequence that we desire.
Next: Jack of All Trades
Last changed by: A.Honegger,