|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Hemagglutinin
Exploring the Structure
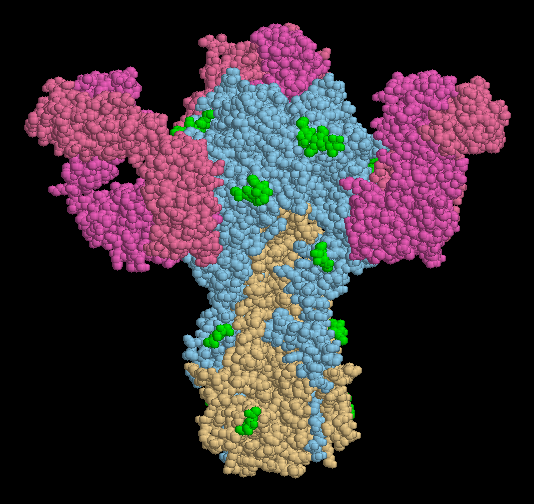
Antibodies are our first line of defense against influenza virus. PDB entry 1qfu shows how one antibody attacks hemagglutinin, blocking it so that it cannot bind to cell surfaces. The structure includes hemagglutinin, shown in blue and orange, and three copies of a Fab fragment of antibody (Fab fragments are one arm from the Y-shaped antibody). Of course, viruses find ways of evading attack by antibodies, creating new strains that infect us each year. One way is to mutate the location of carbohydrate chains on the hemagglutinin surface. Several of these carbohydrates are shown in green here. If the virus adds a new carbohydrate at the location that the antibody binds, the antibody will no longer be effective.
This picture was created with RasMol. You can create similar pictures by clicking on the PDB accession codes and picking one of the options under Display Molecule. Be sure to use the biological unit file when you are looking at this hemagglutinin-antibody structure!
A list of 'hemagglutinin' related entries in the PDB as determined by a keyword search on March 30, 2006 is available. For more information on hemagglutinin, click here.
Next: Interaktive 3D-Animation
Previous Hemagglutinin in Action
Last changed by: A.Honegger,