|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Self-splicing RNA
Exploring the Structure
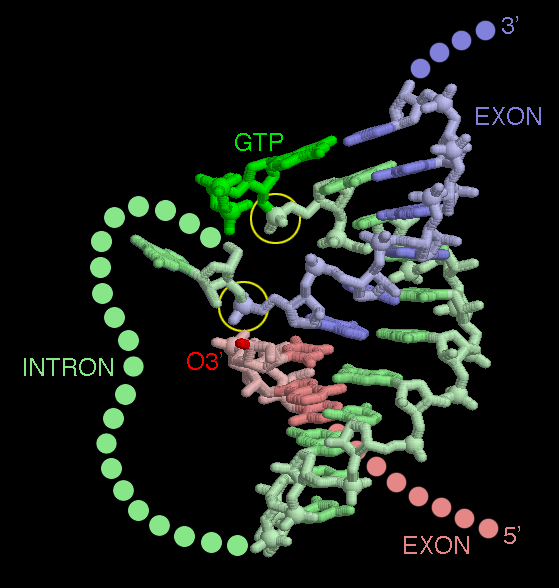
The self-splicing RNA in PDB entry 1u6b has been caught in the middle of its splicing reaction. In this picture, the two exons are shown in red and blue, and part of the intron is shown in green. The rest of the intron (shown on the first page of this Molecule of the Month) is omitted. The reaction starts with one continuous piece of RNA. Imagine that the O3' end of the red exon is attached to the phosphate group circled at the top. Then, if you follow the chain, you go from red exon, through the green intron, and out the blue exon. The structure shows the molecule after the first cleavage has been made. The GTP has broken the chain between the intron and the exon in red, leaving the GTP attached to one end of the intron. At this point, the O3' atom of the red exon is perfectly placed to attack the phosphate at the end of the blue exon (lower circle). When that bond is made, the green intron will be released, and the red and blue exons will be spliced together. Notice that the short portion of the intron shown here, termed the "guide sequence," aligns the two exons perfectly next to one another.This picture was created with RasMol. You can create similar pictures by clicking the accession codes here and picking one of the options under View Structure. When you look at this structure yourself, take a moment to play with coloring of chains, since the overall structure can be confusing. The exon colored red is chain D and exon colored blue is residue numbers 1-6 in chain C. Chain A is a protein that was used to assist in the crystallization, and everything else (chain B and the remainder of chain C) is the intron.
A list of all Self-splicing RNA entries in the PDB as determined by a FASTA search on April 27, 2005 is available here. For more information on ribozymes, click here.
Next: Interaktive 3D-Animation
Previous: The Smallest Ribozyme
Last changed by: A.Honegger,