|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Aminoacyl-tRNA Synthetase
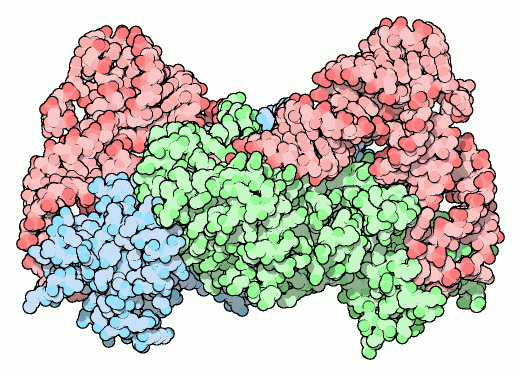
When a ribosome pairs a "CGC" tRNA with "GCG" codon, it expects to find an alanine carried by the tRNA. It has no way of checking; each tRNA is matched with its amino acid long before it reaches the ribosome. The match is made by a collection of remarkable enzymes, the aminoacyl-tRNA synthetases. These enzymes charge each tRNA with the proper amino acid, thus allowing each tRNA to make the proper translation from the genetic code of DNA into the amino acid code of proteins.
Twenty Flavors
Most cells make twenty different aminoacyl-tRNA synthetases, one for each type of amino acid. These twenty enzymes are widely different, each optimized for function with its own particular amino acid and the set of tRNA molecules appropriate to that amino acid.
The one shown here, which charges aspartic acid onto the proper tRNA (entry 1asz), is a dimer of two identical subunits (colored blue and green, the two tRNA molecules are colored red). Others are small monomers or large monomers, or dimers, or even tetramers of one or more different types of subunits. Some have wildly exotic shapes, such as the serine enzyme (entry 1set). The structures of nearly all of these different enzymes are available in the PDB.
Finding the Proper Mate
As you might expect, many of these enzymes recognize their tRNA molecules using the anticodon. But this may not be possible in some cases. Take serine, for instance. Six different codons specify serine, so seryl-tRNA synthetase must recognize six tRNA molecules with six different anticodons, including AGA and GCU, which are entirely different from one another. So, tRNA molecules are also recognized using segments on the acceptor end and bases elsewhere in the molecule. One base in particular, number 73 in the sequence, seems to play a pivotal role in many cases, and has been termed the discriminator base. In other cases, however, it is completely ignored. Note also that each enzyme must recognize its own tRNA molecules, but at the same time, it must not bind to any of the other ones. So, each tRNA has a set of positive interactions that match up the proper tRNA with the proper enzyme, and a set of negative interactions that block binding of improper pairs. For instance, the aspartyl-tRNA synthetase shown here (entry 1asz) recognizes the discriminator base and 4 bases around the anticodon. But, one other base, guanine 37, is not used in binding, but must be methylated to ensure that the tRNA does not bind improperly to the arginyl-tRNA synthetase.
Surprises from Genome Analyses
Recent analyses of entire genomes revealed a big surprise: some organisms don't have genes for all twenty aminoacyl-tRNA synthetases. They do, however, use all twenty amino acids to construct their proteins. The solution to this paradox revealed, as is often the case in living cells, that more complex mechanisms are used. For instance, some bacteria do not have an enzyme for charging glutamine onto its tRNA. Instead, a single enzyme adds glutamic acid to all of the glutamic acid tRNA molecules and to all of the glutamine tRNA molecules. A second enzyme then converts the glutamic acid into glutamine on the latter tRNA molecules, forming the proper pair.
Last changed by: A.Honegger,