|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
Clathrin
Exploring the Structure
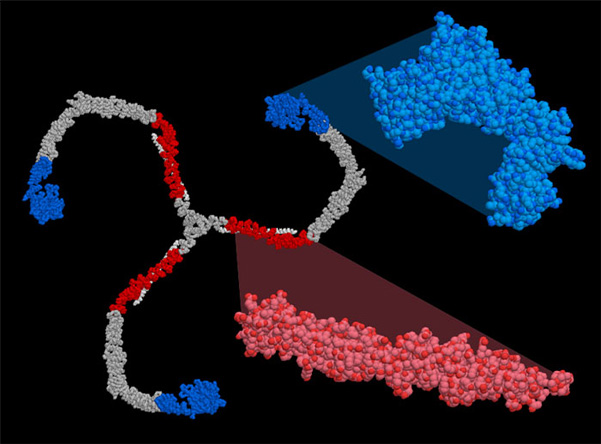
Like the ribosome, the large size and flexibility of clathrin makes it difficult to crystallize, so a variety of lower-resolution visualization technologies were employed to determine its structure. In the case of PDB entry 1xi4, researchers started with two pieces of information. First, they used cryoelectron microscopy to average thousands of electron microscope photographs of individual clathrin cages after freezing them in amorphous ice. Then, they fit the atomic structures of two pieces of clathrin, which had been solved by crystallography (PDB entries 1b89 and 1bpo, shown in red and blue on the right), into 3D image. Atomic models for the missing parts were generated by homology modeling, since the amino acid sequence of clathrin is highly repetitive. The structures were fit into the density map and their ends simply tied together in a manner that simultaneously matches the fit with the known amino acid sequence.
This picture was created using RasMol. You can create similar illustrations by clicking on the accession codes here and picking one of the options under Images and Visualization. When you go to look at the whole clathrin cage in PDB entry 1xi4, be sure to download the entire biological unit!
To see the sources of information that we used for writing this Molecule of the Month, click here. A list of 'clathrin' related entries in the PDB as determined by a keyword search on March 30, 2007 is also available.
Next: Jmol Animation
Previous: Special Delivery
Last changed by: A.Honegger,