|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
TATA-Binding Protein
Exploring the Structure
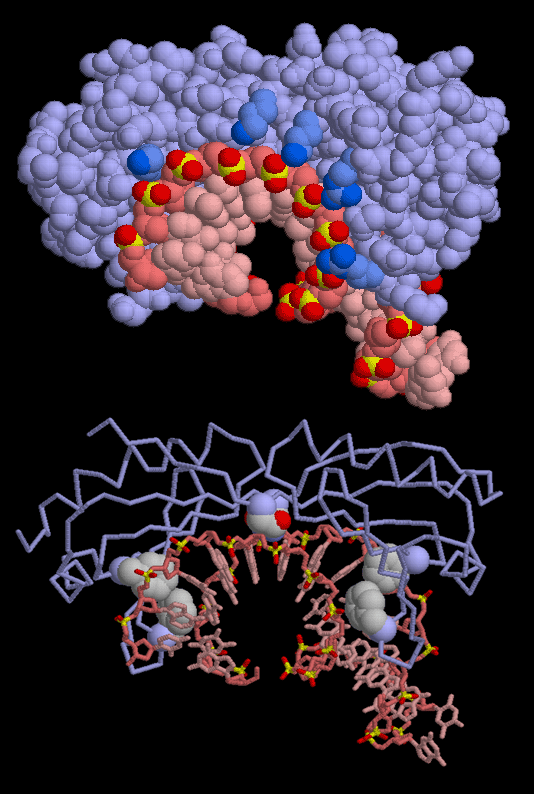
TATA-binding protein uses two types of interactions to recognize and hold the TATA sequence, as seen in this structure from PDB entry 1ytb. First, as shown at the top, it has a string of lysine and arginine amino acids (colored dark blue) that interact with the phosphate groups of the DNA (colored bright yellow and red). This glues the protein to the DNA. Second, the protein uses specially-placed amino acids to interact with DNA bases. As shown in the lower picture, four phenylalanine amino acids jam into the DNA minor groove and form the kinks that bend the DNA. There are also two symmetrical asparagine amino acids that form hydrogen bonds at the very center. The combination of the unusual flexibility of TATA DNA sequences and these specific hydrogen bonds allows TATA-binding protein to recognize the proper sequence.As you are looking at these structures yourself, notice that TATA-binding protein, even though it is composed of a single protein chain, is composed of two symmetrical halves. This symmetry is easily seen in the two pairs of phenylalanines and the two asparagines shown in the lower figure. It is thought that an ancient gene duplication created this protein by combining two copies of the same gene. For more information on TATA-binding protein from a genomics perspective, visit the Protein of the Month at the European Bioinformatics Institute.
These pictures were created with RasMol. You can create similar pictures by clicking on the accession codes here and picking one of the options under View Structure. The phenylalanines shown above are numbers 99, 116, 190, and 207, and the asparagines are numbers 69 and 159.
A list of TATA-binding protein related entries in the PDB as determined by a FASTA search on June 28, 2005 is available here. For more information on TATA-binding protein, click here.
Next: Interaktive 3D-Animation
Previous: Helpers
Last changed by: A.Honegger,