|
Inhaltsübersicht | Nanomaschinen | Moleküle | Programme | Kurse | Fun | Links |
||
| > |
HIV Protease
Exploring the Structure
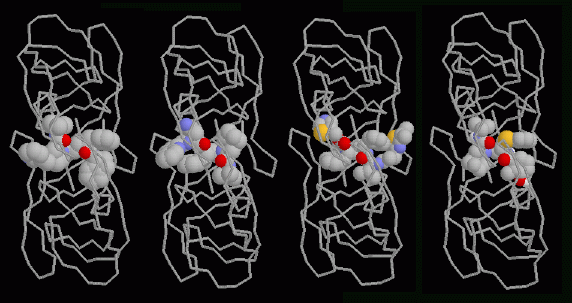
Four drugs that attack HIV-1 protease are currently being used to treat people infected with the virus. Structures of all four of these drugs bound to HIV-1 protease are available at the PDB. In the illustration, the enzyme is displayed as a ribbon that follows the two protein chains and the drugs are shown as spacefilling models. The view is from the top--notice how the flaps cover the top of the drug molecules. From left to right, the drugs are Indinavir (PDB entry 1hsg), Saquinavir (PDB entry 1hxb), Ritonavir (PDB entry 1hxw), and Nelfinavir (PDB entry 1ohr). Notice how similar these drugs are. They all have carbon-rich groups arrayed along either side, interacting with the sides of the active site tunnel. They each have two oxygen atoms at the center, pointing towards us in the illustration, that interact with a special water molecule that is normally trapped under the flaps (not shown here). The drugs all mimic a protein chain, binding to the enzyme like protein chains do. But they are more stable than a protein chain. HIV-1 protease cannot cleave them, so they stay lodged in the active site, blocking the normal function of the enzyme.This illustration was created with RasMol. You can create similar pictures of these drug complexes by clicking on the PDB accession codes above, and then picking "View Structure."
Next: Interaktive 3D-Animation
Previous: A Small But Effective Enzyme
Last changed by: A.Honegger,